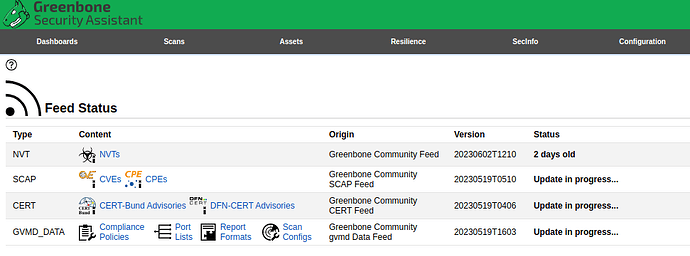
Looks like greenbone/vulnerability-tests image version 5958053f7549 is corrupted.
I trying to run communirty version, but in the gvmd container logs I see the repeated error message:
md manage: INFO:2023-05-19 10h48.26 UTC:86: SCAP database does not exist (yet), skipping CERT severity score update
md manage: INFO:2023-05-19 10h48.26 UTC:86: sync_cert: Updating CERT info succeeded.
noname.xml:670532: parser error : internal error: Huge input lookup
src=g&hsa_net=adwords&hsa_mt=&hsa_ver=3&hsa_ad=111046699381&
^
noname.xml:670532: parser error : attributes construct error
^
noname.xml:670532: parser error : Couldn't find end of Start Tag reference line 670532
^
noname.xml:670532: parser error : EndTag: '</' not found
^
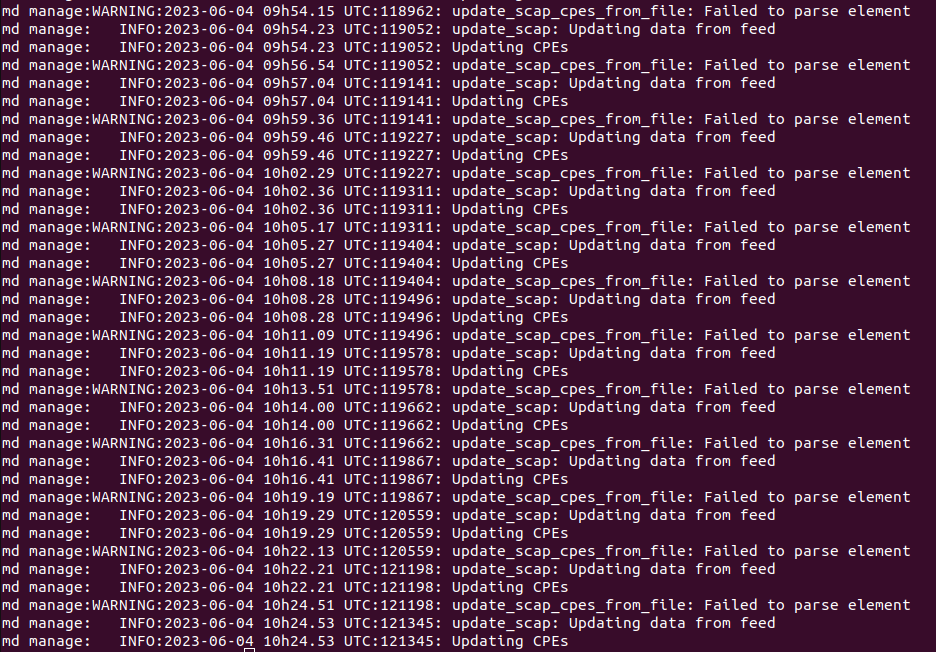
md manage:WARNING:2023-05-19 10h49.42 UTC:84: update_scap_cpes_from_file: Failed to parse element
md manage:WARNING:2023-05-19 10h49.52 UTC:132: update_scap: No SCAP db present, rebuilding SCAP db from scratch
md manage: INFO:2023-05-19 10h49.52 UTC:134: OSP service has different VT status (version 202305191005) from database (version (null), 0 VTs). Starting update ...
md manage: INFO:2023-05-19 10h49.52 UTC:132: update_scap: Updating data from feed
md manage: INFO:2023-05-19 10h49.52 UTC:132: Updating CPEs
noname.xml:670532: parser error : internal error: Huge input lookup
src=g&hsa_net=adwords&hsa_mt=&hsa_ver=3&hsa_ad=111046699381&
^
noname.xml:670532: parser error : attributes construct error
^
noname.xml:670532: parser error : Couldn't find end of Start Tag reference line 670532
^
noname.xml:670532: parser error : EndTag: '</' not found
^
md manage:WARNING:2023-05-19 10h51.53 UTC:132: update_scap_cpes_from_file: Failed to parse element
md manage: INFO:2023-05-19 11h02.00 UTC:134: Updating VTs in database ... 128431 new VTs, 0 changed VTs
md manage: INFO:2023-05-19 11h02.03 UTC:134: Updating VTs in database ... done (128431 VTs).
md manage:WARNING:2023-05-19 11h02.05 UTC:406: update_scap: No SCAP db present, rebuilding SCAP db from scratch
md manage: INFO:2023-05-19 11h02.06 UTC:406: update_scap: Updating data from feed
md manage: INFO:2023-05-19 11h02.06 UTC:406: Updating CPEs
noname.xml:670532: parser error : internal error: Huge input lookup
src=g&hsa_net=adwords&hsa_mt=&hsa_ver=3&hsa_ad=111046699381&
^
noname.xml:670532: parser error : attributes construct error
^
noname.xml:670532: parser error : Couldn't find end of Start Tag reference line 670532
^
noname.xml:670532: parser error : EndTag: '</' not found
^
md manage:WARNING:2023-05-19 11h04.07 UTC:406: update_scap_cpes_from_file: Failed to parse element
md manage:WARNING:2023-05-19 11h04.16 UTC:454: update_scap: No SCAP db present, rebuilding SCAP db from scratch
md manage: INFO:2023-05-19 11h04.17 UTC:454: update_scap: Updating data from feed
md manage: INFO:2023-05-19 11h04.17 UTC:454: Updating CPEs
noname.xml:670532: parser error : internal error: Huge input lookup
src=g&hsa_net=adwords&hsa_mt=&hsa_ver=3&hsa_ad=111046699381&
^
noname.xml:670532: parser error : attributes construct error
^
noname.xml:670532: parser error : Couldn't find end of Start Tag reference line 670532
^
noname.xml:670532: parser error : EndTag: '</' not found
Could you please check if automation is build such image are working corretly?