I have installed OpenVAS/GVM for ubuntu using the following docs - Greenbone Community Containers - Greenbone Community Documentation
Now I need to interact with GVM using python. Previously I installed gvm manually using sudo apt install gvm on kali linux and interacted with it successfully using python-gvm through UnixSocketConnection. But this approach is not working for ubuntu system.
Manual installation on ubuntu was not successful that is why I used greenbone community containers and everything is working fine. I just need to figure out how do I interact with GVM in python using this approach.
I would really appreciate and be grateful for any help and guidance.
Thank you.
bricks
February 18, 2025, 7:20am
2
Hi, please take a look at
and
1 Like
Thanks for the documentation.
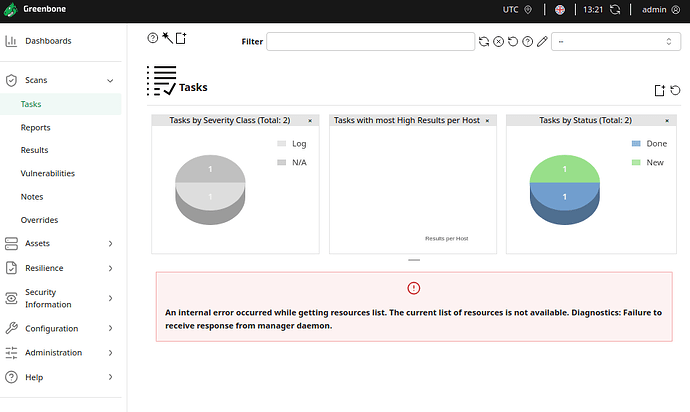
I followed the second part of exposing the Unix socket. This solution worked partially. I am able to start a scan using python but I cant fetch the status. On the GUI I am getting internal server error.
Also, I would really appreciate if you can confirm that I have made correct changes. Here is the part of the compose file.
gvmd:registry.community.greenbone.net/community/gvmd:stable
gvmd_data_vol:/var/lib/gvm
scap_data_vol:/var/lib/gvm/scap-data/
cert_data_vol:/var/lib/gvm/cert-data
data_objects_vol:/var/lib/gvm/data-objects/gvmd
vt_data_vol:/var/lib/openvas/plugins
psql_data_vol:/var/lib/postgresql
/tmp/gvm/gvmd:/run/gvmd
ospd_openvas_socket_vol:/run/ospd
psql_socket_vol:/var/run/postgresql
gsa:registry.community.greenbone.net/community/gsa:stable
127.0.0.1:9392:80
/tmp/gvm/gvmd:/run/gvmd
gvmd
Thank you.
bricks
February 18, 2025, 12:24pm
4
If the management daemon has an error please take a look at the logs to identify the error. Otherwise we can’t offer any help.
These are the some of the error logs:
gmp:UNKNOWN:2025-02-18 08h16.58 utc:614: handle_get_tasks: GET_TASKS: error finding task target, aborting
notus-data-1 | products/suse_linux_enterprise_workstation_extension_12.notus
Is there any I can reinstall the database/delete existing targets or scans because my GUI wont let me. Or do I need to setup the images again.
bricks
February 21, 2025, 4:10pm
6
I haven’t seen such an issue before. I suppose that something is broken in your setup/database.
You could delete the docker volume for the postgres database to remove all stored data. Additionally take a look at the following link for starting from scratch completely